6 Lab 6: Biochemical Tests and Dichotomous Keys
Lab: Biochemical Tests
Lab Objectives
After completing this lab, the student should be able to:
- Use aseptic technique to inoculate biochemical media.
- Classify microbes based on their biochemical results.
- Apply the use of pH indicators to determine metabolic processes by or within a cell.
- Provided a dichotomous key, identify a bacterial unknown.
Microbial identification is key to the clinical sciences. Based on the identification of a causative agent of disease, antimicrobial treatment can be selected that targets the specific pathogen. As you have already experienced with previous labs, microbial identification requires more than just microscopic observations. While staining and microscopy can be a great place to start when identify microbes, it doesn’t provide genus or genus/ species level identification.
Most identification schemes combine Gram reaction and biochemical testing, which typically involves several different tests to narrow down the list possible organisms. There are three main systems that are used within the clinical environment that serve as biochemical testing:
- Biolog system (Biolog)
- API20-e system (bioMerieux)
- EnteroPluri Test (BD Biosciences)
Both the API system and the Biolog system require the generation of a metabolic “footprint” that is then entered into a computer database which will give a genus species level identification. It should be noted that both the Biolog and API systems are quite expensive so are cost prohibitive for many smaller clinics and hospitals thus necessitating that samples be sent to larger medical centers or clinical testing services such as LabCorp®. The Biolog system can identify approximately 3,000 different bacterial and fungal species where as the API system is limited to Gram negative Enterobacteriacaea.
A more cost-effective method for unknown identification of Enterobacteriacaea is the EnteroPluri (Entertube) system in which a Gram negative colony, typically isolated from a selective differential medium such as EMB or MAC is inoculated into one tube that contains 15 different biochemical tests. The Enteropluri tube is incubated overnight and color changes in the medium are compared to a manual that will provide genus species identification of the most common enteric pathogens and opportunists. It should be noted that while the EnteroPluri tube is the most cost effective of these three methods, it also has the most limitations in that it uses the fewest number of biochemical tests which can lead to misidentification.
Because these systems dependent upon biochemical testing rather than genetic testing, identification typically takes longer – 24 hours for the Biolog, API20e, and Enteropluri systems compared to 2-4hr for genetic identifications. Both genetic and biochemical methods can be “tricked” by microbial genetic exchange processes such as conjugation, transduction, and transformation in which non-pathogen genetic material may be taken up by the bacterial cell and result in the expression of new genetic traits. In such cases, more robust systems such as the Biolog system provide a higher confidence in genus/ species level identification.
In this lab, you will use data collected from a bacterial unknown and the associated assignment Lab: Dichotomous Key to identify a specific unknown. Because multiple types of media are being used which can be costly, data will be supplied to you via Canvas message for your unknown organism. For the Results portion of this lab, you will answer questions about the different biochemical tests.
Method (Lab@Home)
For this lab you will need:
Lab: Dichotomous Keys
Biochemicals Lab Lecture
Lab: Biochemicals
Biochemical Data sets 1 and 2
Analysis of provided data
Watch the Lab: Biochemical Tests video.
Using the two data sets (Biochemical data set 1 and Biochemical Data set 2), complete the data tables in the Results section.
Results
Table 1. Results from Carbohydrate Fermentation Tests. For Phenol Red tests record broth color and presence/ absence of bubbles in Durham tube and color change (A = acid; G= gas; AG = acid and gas; K = alkaline reaction; Neg = no reaction). For the OF glucose test, record deep color for the open and closed (with mineral oil) tubes. For MR, record color changes (if any) after the addition of reagents.
|
|
Phenol Red Fermentation
|
OF Glucose
|
MR
|
|||
|
Organism
|
Glucose
|
Lactose
|
Sucrose
|
Open
|
Closed
|
Color after Reagent
|
|
E. aerogenes
|
|
|
|
|
|
|
|
E. coli
|
|
|
|
|
|
|
|
P. vulgaris
|
|
|
|
|
|
|
|
S. aureus
|
|
|
|
|
|
|
|
Negative
|
|
|
|
|
|
|
Table 2. Results of Urea Hydrolysis Test.
|
Organism
|
Agar Color
|
Urease (+/-)
|
|
E. coli
|
|
|
|
P. vulgaris
|
|
|
|
Negative control
|
|
|
Table 3. Results of Citrate Test.
|
Organism
|
Growth (Y/N)
|
Agar Color
|
Citrase (+/-)
|
|
C. freundii
|
|
|
|
|
E. coli
|
|
|
|
|
Negative control
|
|
|
|
Table 4. Results of Catalase Test.
|
Organism
|
Bubbles (+/-)
|
Catalase (+/-)
|
|
E. faecalis
|
|
|
|
S. aureus
|
|
|
|
Negative control.
|
|
|
Table 5. Results of SIM Test.
|
|
Sulfide Test
|
Indole Test
|
Motility
|
|
|
Organism
|
Agar Color
|
Agar Color
|
Tryptophanase (+/-)
|
(+/-)
|
|
C. freundii
|
|
|
|
|
|
E. aerogenes
|
|
|
|
|
|
E. coli
|
|
|
|
|
|
P. vulgaris
|
|
|
|
|
|
Negative control
|
|
|
|
|
Table 6. Results of Oxidase Test.
|
Organism
|
Color
|
Oxidase (+/-)
|
|
E. coli
|
|
|
|
P. vulgaris
|
|
|
|
P. aeruginosa
|
|
|
| Negative Control |
|
|
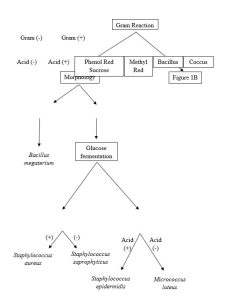
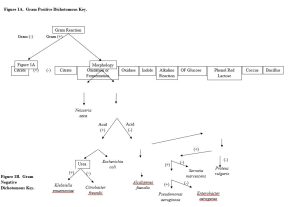
Lab: Dichotomous Keys
Objectives
After completing this lab, students will be able to:
- Provided a dichotomous key, identify a bacterial unknown.
Introduction
These dichotomous keys will be used to identify microbes isolated from human samples or provided unknown data sets.
Method (Lab@Home)
For this lab you will need:
Data set (sent to your Canvas email)
Protocol (with dichotomous key)
Instructions for data collection
Complete the two tables below by adding data for each (Observations and Interpretations) for each biochemical test used. If you did not use a test then mark the “Observation” and “Interpretation” fields as NA or with a dash (—).
1. Data collected for the Unknown Bacterium. Unknown #: __________
|
Biochemical Test
|
Observation
|
Interpretation
|
|
Gram Reaction
|
|
|
|
Morphology
|
|
|
|
Arrangement
|
|
|
|
Phenol Red Glucose
|
|
|
|
Phenol Red Lactose
|
|
|
|
Phenol Red Sucrose
|
|
|
|
OF Glucose
|
|
|
|
Methyl Red
|
|
|
|
Urea
|
|
|
|
Citrate
|
|
|
|
Catalase
|
|
|
|
SIM
|
|
|
|
Oxidase
|
|
|